Nested Slice Sampling
ICLR 2025 Workshop Frontiers in Probabilistic Inference
Presented at ICLR 2025 Workshop: Frontiers in Probabilistic Inference | OpenReview | Poster (PDF)
Alongside co-authors Namu Kroupa and Will Handley, we present an updated, GPU-compatible nested sampling implementation.
Key result: A high-performance nested sampling implementation in JAX/blackJAX achieving significant GPU speedups while maintaining competitive accuracy with state-of-the-art SMC methods.
Nested Sampling as Particle Monte Carlo
Nested sampling estimates the normalizing constant (evidence) associated with a target distribution:
$$Z(\beta) = \int e^{-\beta E(x)} \Pi(x) dx$$
Unlike standard annealing approaches that construct a single temperature path from the prior \( \Pi(x) \) to the posterior \( P(x) \), nested sampling constructs a unique path through constrained regions of prior mass. These constraints arise from progressively increasing likelihood thresholds.
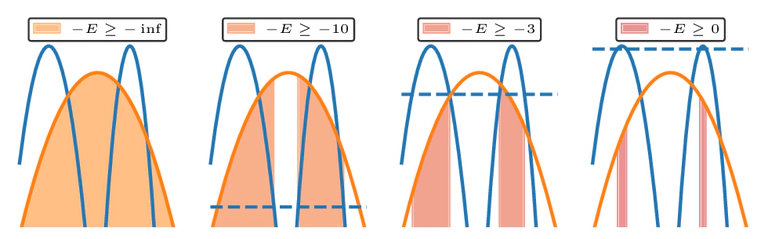
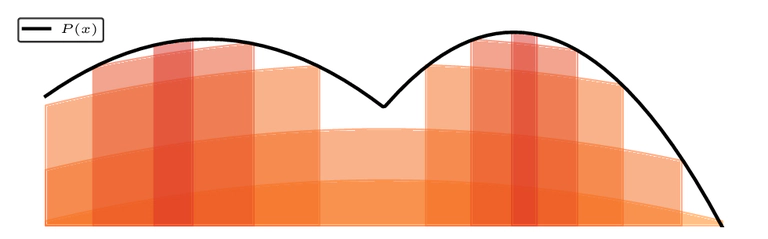
Vectorization and GPU Acceleration
Our implementation, Nested Slice Sampling (NSS), achieves high GPU utilization for parallel particle updates. Batch updates trade modest evidence accuracy for significant walltime speedups—updating particle subsets yields order-of-magnitude improvements on GPUs, with a manageable cost in evidence estimation accuracy.
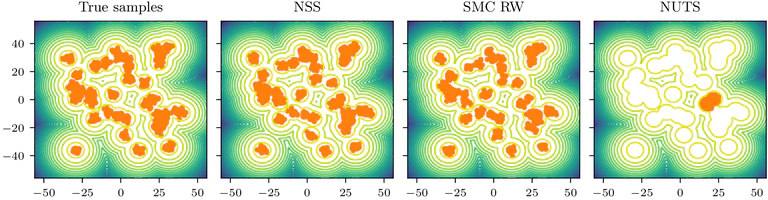
NSS compares favorably with SMC for state-of-the-art performance on challenging inference tasks. The probabilistic volume estimates produced by NSS provide unique advantages: evidence estimation with built-in error quantification.
Takeaways
- High-Performance Nested Sampling in JAX/blackJAX — Open source implementation integrated with the blackJAX ecosystem
- Massively Parallel Execution — Significant speedups on GPUs/TPUs via vectorization
- Runtime Competitive with State-of-the-Art SMC — Matches or exceeds leading population MC methods on benchmark tasks
- Robust Bayesian Evidence Estimation — Excels on complex geometries, scaling efficiently to \(\mathcal{O}(100)\) dimensions
Code available at handley-lab/blackjax. See also the Nested Sampling Book for further documentation.
